NMF for digits feature extraction¶
Non-negative matrix factorization (NMF) with sparseness enforced in the components, in comparison with PCA for feature extraction.
Python source code: plot_nmf.py
print __doc__
from time import time
import logging
import pylab as pl
from scikits.learn.decomposition import RandomizedPCA, NMF
from scikits.learn import datasets
# Display progress logs on stdout
logging.basicConfig(level=logging.INFO,
format='%(asctime)s %(levelname)s %(message)s')
digits = datasets.load_digits()
# reshape the data using the traditional (n_samples, n_features) shape
n_samples = len(digits.images)
X = digits.images.reshape((n_samples, -1))
n_features = X.shape[1]
n_components = 16
######################################################################
# Compute a PCA (eigendigits) on the digit dataset
print "Extracting the top %d eigendigits from %d images" % (
n_components, X.shape[0])
t0 = time()
pca = RandomizedPCA(n_components=n_components, whiten=True).fit(X)
print "done in %0.3fs" % (time() - t0)
eigendigits = pca.components_
######################################################################
# Compute a NMF on the digit dataset
print "Extracting %d non-negative features from %d images" % (
n_components, X.shape[0])
t0 = time()
nmf = NMF(n_components=n_components, init='nndsvd', beta=5, tol=1e-2,
sparseness="components").fit(X)
print "done in %0.3fs" % (time() - t0)
nmfdigits = nmf.components_
######################################################################
# Plot the results
n_row, n_col = 4, 4
f1 = pl.figure(figsize=(1. * n_col, 1.13 * n_row))
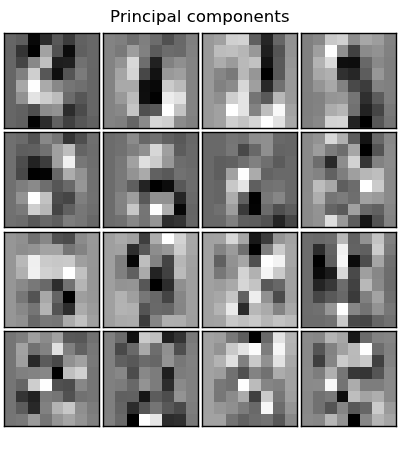
f1.text(.5, .95, 'Principal components', horizontalalignment='center')
for i in range(n_row * n_col):
pl.subplot(n_row, n_col, i + 1)
pl.imshow(eigendigits[i].reshape((8, 8)), cmap=pl.cm.gray,
interpolation='nearest')
pl.xticks(())
pl.yticks(())
pl.subplots_adjust(0.01, 0.05, 0.99, 0.93, 0.04, 0.)
f2 = pl.figure(figsize=(1. * n_col, 1.13 * n_row))
f2.text(.5, .95, 'Non-negative components', horizontalalignment='center')
for i in range(n_row * n_col):
pl.subplot(n_row, n_col, i + 1)
pl.imshow(nmfdigits[i].reshape((8, 8)), cmap=pl.cm.gray,
interpolation='nearest')
pl.xticks(())
pl.yticks(())
pl.subplots_adjust(0.01, 0.05, 0.99, 0.93, 0.04, 0.)
pl.show()